SPIE-AAPM-NCI BreastPathQ: Cancer Cellularity Challenge 2019¶
The SPIE (the international society for optics and photonics), along with the American Association of Physicists in Medicine (AAPM), and the National Cancer Institute (NCI), will conduct a “Grand Challenge” on the development of quantitative biomarkers for the determination of cancer cellularity from whole slide images (WSI) of breast cancer hematoxylin and eosin (H&E) stained pathological slides. As part of the 2019 SPIE Medical Imaging Conference, the BreastPathQ Challenge will provide a unique opportunity for participants to compare their algorithms with those of others from academia, industry, and government in a structured, direct way using the same data sets.
We have introduced a cancer cellularity scoring challenge for tumor burden assessment in breast pathology. Participants will be tasked to develop an automated method for analyzing histology patches extracted from whole slide images and assign a score reflecting cancer cellularity in each. Currently, this task is performed manually and relies upon expert interpretation of complex tissue structures. Furthermore, reproducibility of cancer cellularity scores is a concern in current practice, therefore a fully automated method holds great promise for increasing throughput and reducing inter- and intra-observer variability.
Results¶
Note that the official challenge is now over. This site is for a continuation of the challenge with a public leaderboard. The dates and prizes are shown for historical purposes. The results of the challenge have been released to the public. The histogram of the results are below.
 ¶
¶
Important Dates¶
Prizes¶
A joint computer-aided diagnosis (CAD) and digital pathology session
at the 2019 SPIE Medical Imaging Conference will focus on the
BreastPathQ Challenge; an individual from each of the three
top-performing teams of the challenge will receive a waiver of the
meeting registration fee in order to present their methods during this
session. After the completion of the BreastPathQ challenge, all
participants will be invited to submit their work in a collaborative
journal paper and each participating team will be allowed up to two
co-authorships.
Background¶
Neoadjuvant treatment (NAT) of breast cancer (BCa) is an option for patients with the locally advanced disease. Moreover, tumor response to the therapy provides useful information for patient management. In addition to the treatment effect on tumor size, NAT may alter the tumor cellularity. Tumor size many not decrease, but the overall cellularity may be markedly reduced, making residual tumor cellularity an important factor in assessing response. The pathological examination of the tissue sections after surgery is the gold-standard to estimate the residual tumor and the assessment of cellularity is an important component of tumor burden assessment. Cellularity within the tumor bed is defined as the percentage area of the overall tumor bed that is comprised of tumor cells (invasive or in situ). The most acceptable methods of assessing residual cancer burden in ongoing clinical trials and in clinical practice in some centers follows the algorithm proposed by an international working group. This algorithm takes into account several parameters including tumor cellularity. In the current clinical practice, tumor cellularity is manually estimated by pathologists on hematoxylin and eosin (H&E) stained slides, the quality, and reliability of which might be impaired by inter-observer variability which potentially affects prognostic power assessment in NAT trials.
Dataset¶
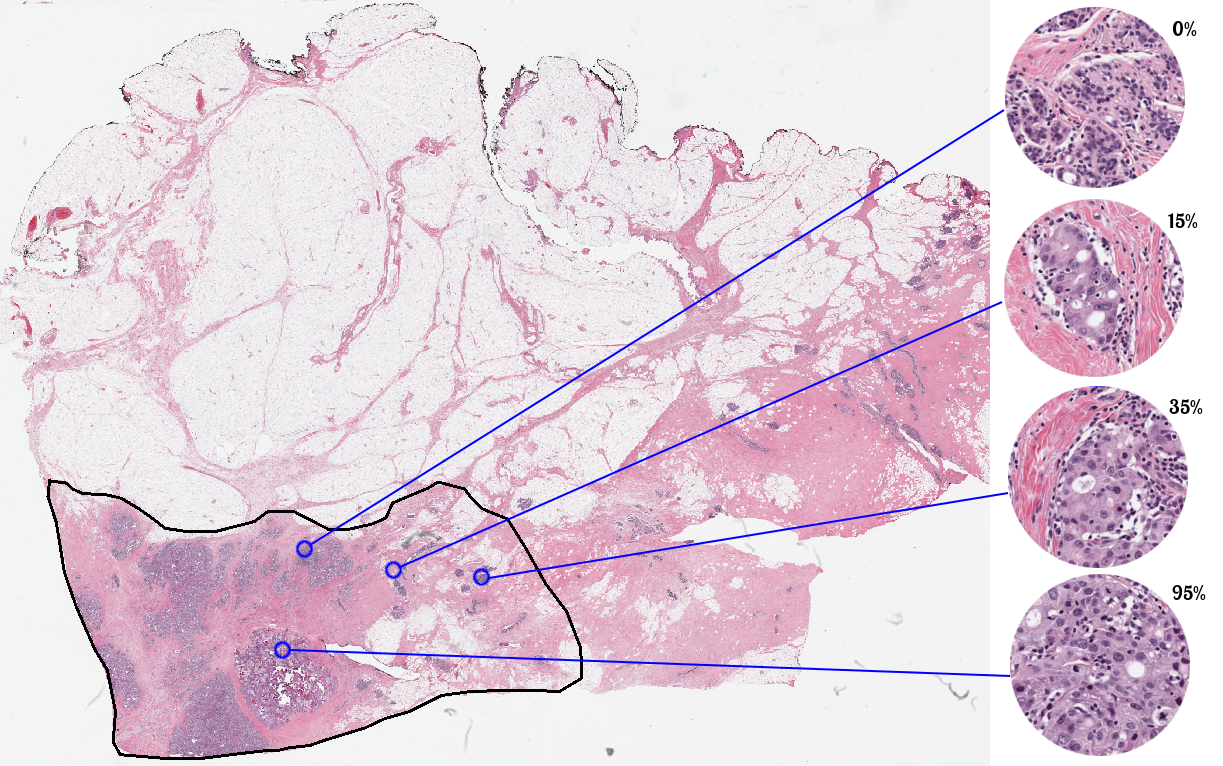
The dataset for this challenge was collected at the Sunnybrook Health Sciences Centre, Toronto. It comprises 96 whole slide images (WSI) which have been stained with haematoxylin and eosin (H&E), a commonly used stain in pathology to highlight cellular and connective tissue. WSIs were extracted from 64 patients with residual invasive breast cancer on resection specimens following neoadjuvant therapy. The specimens were handled according to routine clinical protocols and WSIs were scanned at 20X magnification (0.5 lm/pixel).
In this challenge, you will be provided with a training/validation set of 2,579 patches extracted from the above WSIs. Each patch in the training set has been assigned a tumor cellularity score by 1 expert pathologist, while the test set has reference standards from 2 pathologists. Coordinates for each patch and original WSIs may be provided upon request for additional context. You may choose to use the provided WSIs but it is not a requirement.
The Challenge¶
Participants are required to automatically score each patch on a continuous scale from 0 to 1. Scores must be provided as a floating-point number and must be between 0 and 1 inclusive.
Participants may use the training set cases in any manner they would like for the purpose of training their systems; there will be no restrictions on the use of the data or the advice sought from local experts for training purposes. The test set cases, however, are to be manipulated, processed, and analyzed without human intervention.
Training/Validation Sets The training/validation set has been prepared and contains 2,579 patches extracted from 69 WSIs. Labels will be provided for each training case to be used in algorithm development. Validation labels for 185 patches will originally be withheld for initial validation of developed algorithms but subsequently, the validation labels will be released when the test data becomes available.
Test Set The test set has been prepared in an identical manner as the training set and contains 1,121 patches extracted from 25 WSIs. No labels will be provided for the test set until after the completion of the challenge. You will be asked to run your developed solution on the test set and submit a CSV file per task with your final submission.
Additional Dataset¶
In addition to image patches extracted from whole slide images, we will also provide annotations of lymphocytes, malignant epithelial and normal epithelial cell nuclei in 153 regions-of-interest (ROI) from the same dataset. These annotations may be used in addition to the dataset described above, to distinguish between malignant and healthy structures. Cell nuclei have been marked and x-y co-ordinates stored in an .xml file for each ROI. Usage of this particular data has been described in:
Peikari, M., Salama, S., Nofech-Mozes, S. and Martel, A.L., 2017. Automatic cellularity assessment from post-treated breast surgical specimens. Cytometry Part A, 91(11), pp.1078-1087.
Participants are free to download the training set and, subsequently,
the test set when these datasets become available. It is important to
note that once participants submit their test set cellularity output to
the challenge organizers, they will be considered fully vested in the
Challenge, so that their performance results (without links to the
identity of the participant) will become part of any presentations,
publications, or subsequent analyses derived from the Challenge at the
discretion of the organizers. The truth associated with the test set
cases is expected to be made publicly available after publication of the
BreastPathQ Challenge.
The images for the dataset are available at the Data Sets section on the left of site.
Participation in the BreastPathQ Challenge acknowledges the educational, friendly competition, and community-building nature of this challenge and commits to conduct consistent with this spirit for the advancement of the medical imaging research community. See this link for a discussion of lessons learned from the 2015 LUNGx Challenge, also sponsored by SPIE, AAPM, and NCI.
Challenge Participation¶
To learn more about this challenge, or to participate, please visit
this link.
(Note that the SPIE challenge website is down at this time. Continued
evaluation of the test results will be provided by this site.)
Contact¶
If you have any question regarding this challenge, please contact Kenny Cha at Kenny.Cha@fda.hhs.gov